Phase space for a three-body decay#
Show code cell content
%config InlineBackend.figure_formats = ['svg']
from __future__ import annotations
import os
import warnings
from typing import TYPE_CHECKING
import matplotlib.pyplot as plt
import numpy as np
import sympy as sp
from ampform.sympy import (
UnevaluatedExpression,
create_expression,
implement_doit_method,
make_commutative,
)
from IPython.display import Math
from ipywidgets import FloatSlider, VBox, interactive_output
from matplotlib.patches import Patch
from tensorwaves.function.sympy import create_parametrized_function
if TYPE_CHECKING:
from matplotlib.axis import Axis
from matplotlib.contour import QuadContourSet
from matplotlib.lines import Line2D
warnings.filterwarnings("ignore")
Kinematics for a three-body decay \(0 \to 123\) can be fully described by two Mandelstam variables \(\sigma_1, \sigma_2\), because the third variable \(\sigma_3\) can be expressed in terms \(\sigma_1, \sigma_2\), the mass \(m_0\) of the initial state, and the masses \(m_1, m_2, m_3\) of the final state. As can be seen, the roles of \(\sigma_1, \sigma_2, \sigma_3\) are interchangeable.
Show code cell source
def compute_third_mandelstam(sigma1, sigma2, m0, m1, m2, m3) -> sp.Expr:
return m0**2 + m1**2 + m2**2 + m3**2 - sigma1 - sigma2
m0, m1, m2, m3 = sp.symbols("m:4")
s1, s2, s3 = sp.symbols("sigma1:4")
computed_s3 = compute_third_mandelstam(s1, s2, m0, m1, m2, m3)
Math(Rf"{sp.latex(s3)} = {sp.latex(computed_s3)}")
The phase space is defined by the closed area that satisfies the condition \(\phi(\sigma_1,\sigma_2) \leq 0\), where \(\phi\) is a Kibble function:
Show code cell source
@make_commutative
@implement_doit_method
class Kibble(UnevaluatedExpression):
def __new__(cls, sigma1, sigma2, sigma3, m0, m1, m2, m3, **hints) -> Kibble:
return create_expression(cls, sigma1, sigma2, sigma3, m0, m1, m2, m3, **hints)
def evaluate(self) -> sp.Expr:
sigma1, sigma2, sigma3, m0, m1, m2, m3 = self.args
return Källén(
Källén(sigma2, m2**2, m0**2),
Källén(sigma3, m3**2, m0**2),
Källén(sigma1, m1**2, m0**2),
)
def _latex(self, printer, *args):
sigma1, sigma2, *_ = map(printer._print, self.args)
return Rf"\phi\left({sigma1}, {sigma2}\right)"
@make_commutative
@implement_doit_method
class Källén(UnevaluatedExpression):
def __new__(cls, x, y, z, **hints) -> Källén:
return create_expression(cls, x, y, z, **hints)
def evaluate(self) -> sp.Expr:
x, y, z = self.args
return x**2 + y**2 + z**2 - 2 * x * y - 2 * y * z - 2 * z * x
def _latex(self, printer, *args):
x, y, z = map(printer._print, self.args)
return Rf"\lambda\left({x}, {y}, {z}\right)"
kibble = Kibble(s1, s2, s3, m0, m1, m2, m3)
Math(Rf"{sp.latex(kibble)} = {sp.latex(kibble.doit(deep=False))}")
and \(\lambda\) is the Källén function:
Show code cell source
x, y, z = sp.symbols("x:z")
expr = Källén(x, y, z)
Math(f"{sp.latex(expr)} = {sp.latex(expr.doit())}")
Any distribution over the phase space can now be defined using a two-dimensional grid over a Mandelstam pair \(\sigma_1,\sigma_2\) of choice, with the condition \(\phi(\sigma_1,\sigma_2)<0\) selecting the values that are physically allowed.
Show code cell source
def is_within_phasespace(
sigma1, sigma2, m0, m1, m2, m3, outside_value=sp.nan
) -> sp.Piecewise:
sigma3 = compute_third_mandelstam(sigma1, sigma2, m0, m1, m2, m3)
kibble = Kibble(sigma1, sigma2, sigma3, m0, m1, m2, m3)
return sp.Piecewise(
(1, sp.LessThan(kibble, 0)),
(outside_value, True),
)
is_within_phasespace(s1, s2, m0, m1, m2, m3)
phsp_expr = is_within_phasespace(s1, s2, m0, m1, m2, m3, outside_value=0)
phsp_func = create_parametrized_function(
phsp_expr.doit(),
parameters={m0: 2.2, m1: 0.2, m2: 0.4, m3: 0.4},
backend="numpy",
)
Show code cell source
sliders = {
"m0": FloatSlider(description="m0", max=3, value=2.1, step=0.01),
"m1": FloatSlider(description="m1", max=2, value=0.2, step=0.01),
"m2": FloatSlider(description="m2", max=2, value=0.4, step=0.01),
"m3": FloatSlider(description="m3", max=2, value=0.4, step=0.01),
}
resolution = 300
X, Y = np.meshgrid(
np.linspace(0, 4, num=resolution),
np.linspace(0, 4, num=resolution),
)
data = {"sigma1": X, "sigma2": Y}
sidebar_ratio = 0.15
fig, ((ax1, _), (ax, ax2)) = plt.subplots(
figsize=(7, 7),
ncols=2,
nrows=2,
gridspec_kw={
"height_ratios": [sidebar_ratio, 1],
"width_ratios": [1, sidebar_ratio],
},
)
_.remove()
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.set_xlabel(R"$\sigma_1$")
ax.set_ylabel(R"$\sigma_2$")
ax.set_xticks(range(5))
ax.set_yticks(range(5))
ax1.set_xlim(0, 4)
ax2.set_ylim(0, 4)
for a in [ax1, ax2]:
a.set_xticks([])
a.set_yticks([])
a.axis("off")
fig.tight_layout()
fig.subplots_adjust(wspace=0, hspace=0)
fig.canvas.toolbar_visible = False
fig.canvas.header_visible = False
fig.canvas.footer_visible = False
MESH: QuadContourSet | None = None
PROJECTIONS: tuple[Line2D, Line2D] = None
BOUNDARIES: list[Line2D] | None = None
def plot(**parameters):
draw_boundaries(
parameters["m0"],
parameters["m1"],
parameters["m2"],
parameters["m3"],
)
global MESH, PROJECTIONS
if MESH is not None:
for coll in MESH.collections:
ax.collections.remove(coll)
phsp_func.update_parameters(parameters)
Z = phsp_func(data)
MESH = ax.contour(X, Y, Z, colors="black")
contour = MESH.collections[0]
contour.set_facecolor("lightgray")
x = X[0]
y = Y[:, 0]
Zx = np.nansum(Z, axis=0)
Zy = np.nansum(Z, axis=1)
if PROJECTIONS is None:
PROJECTIONS = (
ax1.plot(x, Zx, c="black", lw=2)[0],
ax2.plot(Zy, y, c="black", lw=2)[0],
)
else:
PROJECTIONS[0].set_data(x, Zx)
PROJECTIONS[1].set_data(Zy, y)
ax1.relim()
ax2.relim()
ax1.autoscale_view(scalex=False)
ax2.autoscale_view(scaley=False)
create_legend(ax)
fig.canvas.draw()
def draw_boundaries(m0, m1, m2, m3) -> None:
global BOUNDARIES
s1_min = (m2 + m3) ** 2
s1_max = (m0 - m1) ** 2
s2_min = (m1 + m3) ** 2
s2_max = (m0 - m2) ** 2
if BOUNDARIES is None:
BOUNDARIES = [
ax.axvline(s1_min, c="red", ls="dotted", label="$(m_2+m_3)^2$"),
ax.axhline(s2_min, c="blue", ls="dotted", label="$(m_1+m_3)^2$"),
ax.axvline(s1_max, c="red", ls="dashed", label="$(m_0-m_1)^2$"),
ax.axhline(s2_max, c="blue", ls="dashed", label="$(m_0-m_2)^2$"),
]
else:
BOUNDARIES[0].set_data(get_line_data(s1_min))
BOUNDARIES[1].set_data(get_line_data(s2_min, horizontal=True))
BOUNDARIES[2].set_data(get_line_data(s1_max))
BOUNDARIES[3].set_data(get_line_data(s2_max, horizontal=True))
def create_legend(ax: Axis):
if ax.get_legend() is not None:
return
label = Rf"${sp.latex(kibble)}\leq0$"
ax.legend(
handles=[
Patch(label=label, ec="black", fc="lightgray"),
*BOUNDARIES,
],
loc="upper right",
facecolor="white",
framealpha=1,
)
def get_line_data(value, horizontal: bool = False) -> np.ndarray:
pair = (value, value)
if horizontal:
return np.array([(0, 1), pair])
return np.array([pair, (0, 1)])
output = interactive_output(plot, controls=sliders)
VBox([output, *sliders.values()])
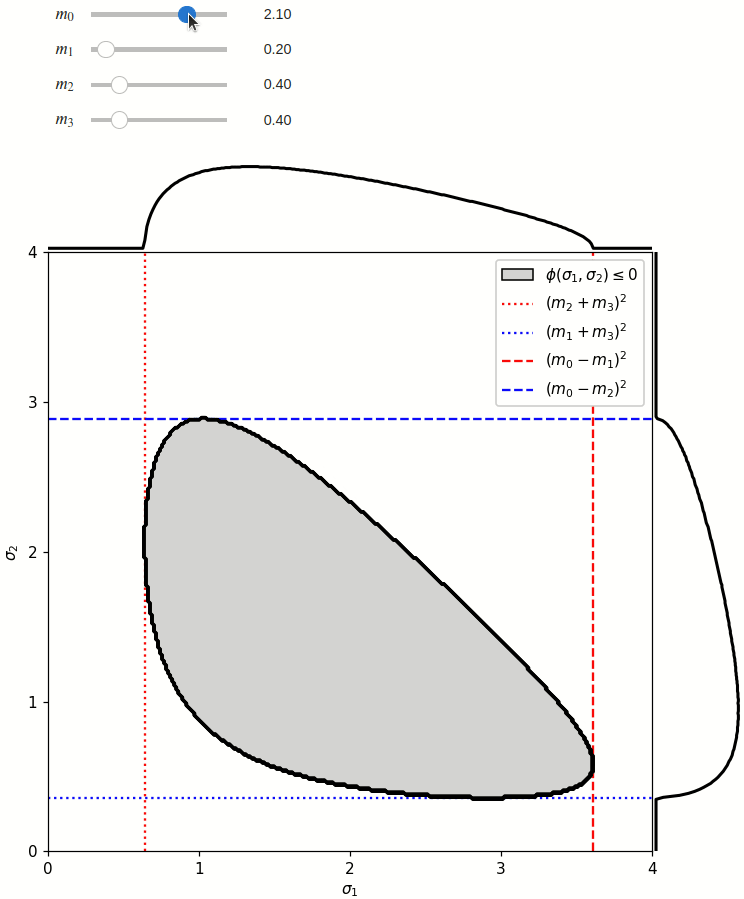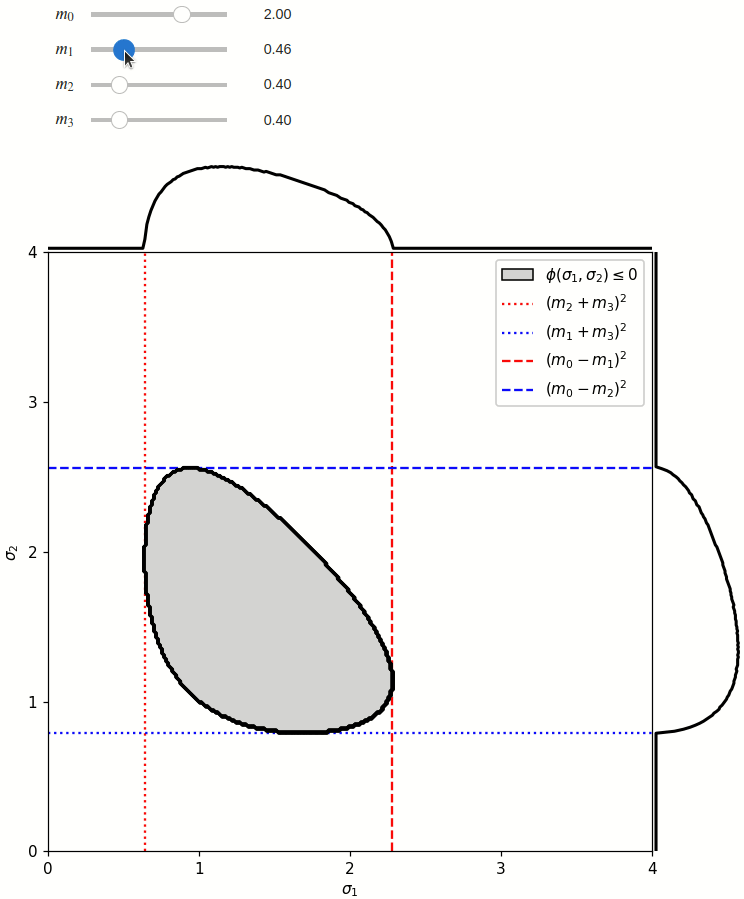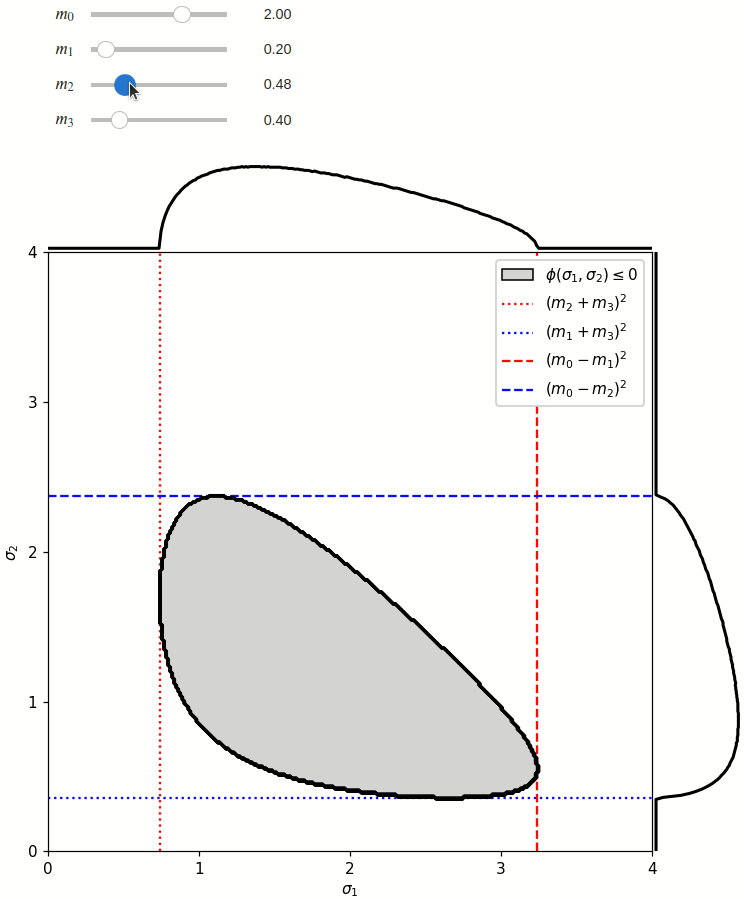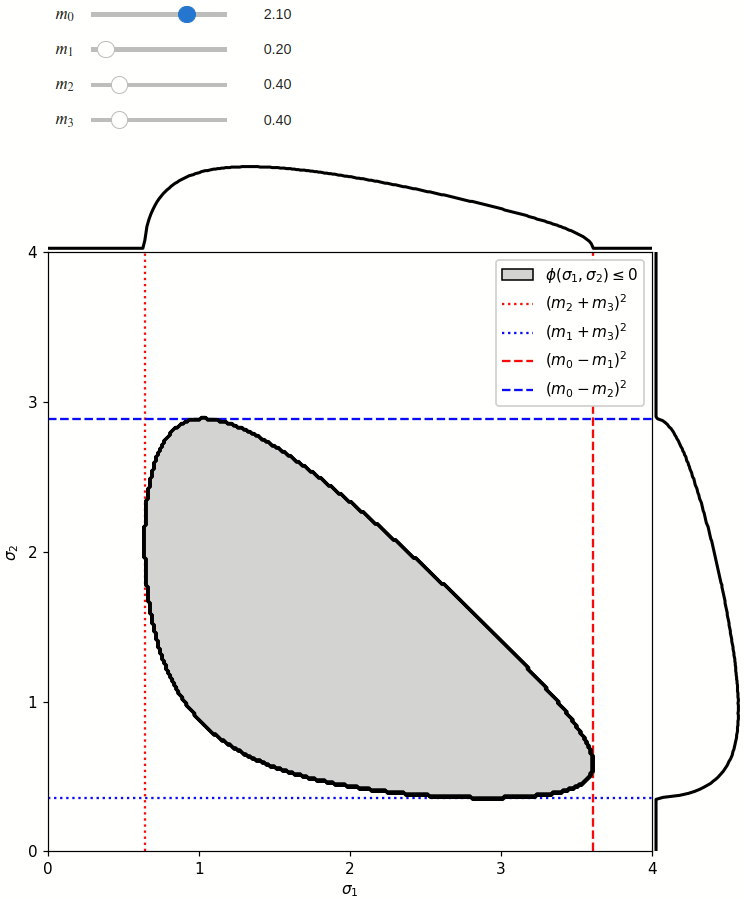
The phase space boundary can be described analytically in terms of \(\sigma_1\) or \(\sigma_2\), in which case there are two solutions:
sol1, sol2 = sp.solve(kibble.doit().subs(s3, computed_s3), s2)
The boundary cannot be parametrized analytically in polar coordinates, but there is a numeric solution. The idea is to solve the condition \(\phi(\sigma_1,\sigma_2)=0\) after the following substitutions:
Show code cell source
T0, T1, T2, T3 = sp.symbols("T0:4")
r, theta = sp.symbols("r theta", nonnegative=True)
substitutions = {
s1: (m2 + m3) ** 2 + T1,
s2: (m1 + m3) ** 2 + T2,
s3: (m1 + m2) ** 2 + T3,
T1: T0 / 3 - r * sp.cos(theta),
T2: T0 / 3 - r * sp.cos(theta + 2 * sp.pi / 3),
T3: T0 / 3 - r * sp.cos(theta + 4 * sp.pi / 3),
T0: (
m0**2 + m1**2 + m2**2 + m3**2 - (m2 + m3) ** 2 - (m1 + m3) ** 2 - (m1 + m2) ** 2
),
}
For every value of \(\theta \in [0, 2\pi)\), the value of \(r\) can now be found by solving the condition \(\phi(r, \theta)=0\). Note that \(\phi(r, \theta)\) is a cubic polynomial of \(r\). For instance, if we take \(m_0=5, m_1=2, m_{2,3}=1\):
Show code cell source
phi_r = (
kibble.doit()
.subs(substitutions) # substitute sigmas
.subs(substitutions) # substitute T123
.subs(substitutions) # substitute T0
.subs({m0: 5, m1: 2, m2: 1, m3: 1})
.simplify()
.collect(r)
)
The lowest value of \(r\) that satisfies \(\phi(r,\theta)=0\) defines the phase space boundary.



