Lecture 12 – γ p [solution]#
Invariant masses of particle pairs and Dalitz plots for another reaction with three particles in the final state \(\gamma p\rightarrow \pi^+\pi^-p\).
Prepare the notebook with the preambles for the inclusion of pandas, numpy and matplotlib.pyplot:
/home/runner/work/strong2020-salamanca/strong2020-salamanca/docs/lecture12a-gammap-solution/.venv/lib/python3.13/site-packages/gdown/cached_download.py:102: FutureWarning: md5 is deprecated in favor of hash. Please use hash='md5:xxx...' instead.
warnings.warn(
| p1x | p1y | p1z | E1 | p2x | p2y | p2z | E2 | p3x | p3y | p3z | E3 | pgamma | helGamma | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.517085 | 0.161989 | 0.731173 | 0.920732 | -0.254367 | 0.082889 | 0.495509 | 0.580189 | -0.259266 | -0.265152 | 0.445175 | 1.10275 | 1.657800 | -1 | |
| 1 | -0.216852 | -0.245333 | 0.107538 | 0.371878 | -0.183380 | 0.191897 | 0.145128 | 0.333213 | 0.389750 | 0.112110 | 0.790133 | 1.29195 | 1.044790 | -1 | |
| 2 | 0.197931 | 0.071432 | -0.010077 | 0.252778 | -0.111780 | -0.360482 | 0.367842 | 0.545221 | -0.090228 | 0.302313 | 0.724911 | 1.22694 | 1.095470 | -1 | |
| 3 | -0.505371 | -0.163949 | 0.450407 | 0.710395 | -0.141057 | 0.282404 | 1.186530 | 1.235730 | 0.679497 | -0.112014 | 0.729706 | 1.37371 | 2.388690 | -1 | |
| 4 | 0.260706 | -0.330303 | 0.385968 | 0.587839 | 0.163863 | 0.354007 | 0.286983 | 0.504031 | -0.490385 | -0.019313 | 0.879568 | 1.37653 | 1.454880 | 1 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 701969 | 0.128575 | -0.018500 | 0.143663 | 0.238807 | 0.378718 | -0.051974 | 0.461311 | 0.615185 | -0.497148 | 0.087386 | 0.483961 | 1.17020 | 1.083930 | -1 | |
| 701970 | -0.262395 | -0.235015 | 0.024120 | 0.379712 | 0.330904 | -0.040001 | 0.751630 | 0.834003 | -0.095422 | 0.256882 | 0.695036 | 1.19938 | 1.492470 | 1 | |
| 701971 | -0.068621 | 0.111528 | -0.032843 | 0.194273 | -0.274412 | -0.101159 | 0.115541 | 0.344094 | 0.325894 | -0.006340 | 0.781223 | 1.26369 | 0.862625 | -1 | |
| 701972 | 0.297759 | -0.100136 | 0.766630 | 0.840194 | 0.034110 | 0.110721 | 0.409582 | 0.447991 | -0.472764 | -0.012095 | 0.440591 | 1.13935 | 2.200460 | -1 | |
| 701973 | 0.169067 | 0.426984 | 0.254915 | 0.543504 | -0.122702 | 0.017682 | -0.012514 | 0.187193 | -0.074462 | -0.430331 | 0.953283 | 1.40707 | 1.587060 | -1 |
701974 rows × 15 columns
The columns headers present some trailing blanks, that must be dropped to be able to use correctly the DataFormat structure (if not, they deliver an error message). To do so, the
str.strip()function must be used beforehand to reformat the column shape.
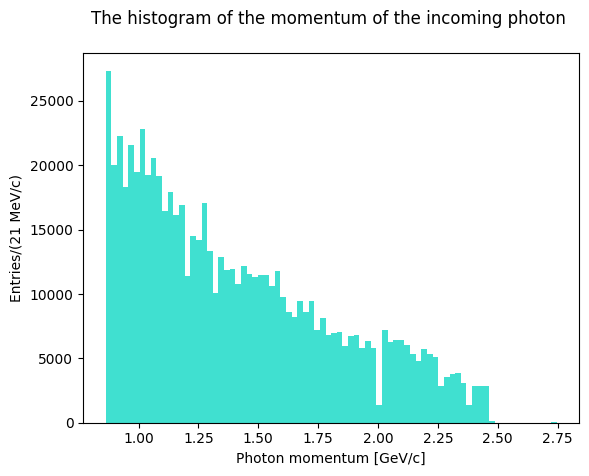
Inspect the data file and format. The file contains the 4-momenta of the particles of the reaction \(\gamma p \rightarrow \pi^+\pi^- p\), the last column corresponds to the 3rd component of the photon momentum (up to 2.5 GeV/c), which travels along the \(z\) axis. Since the target was made of HD, to select the data interacting on protons only a cut on the missing momentum of the reaction was made.
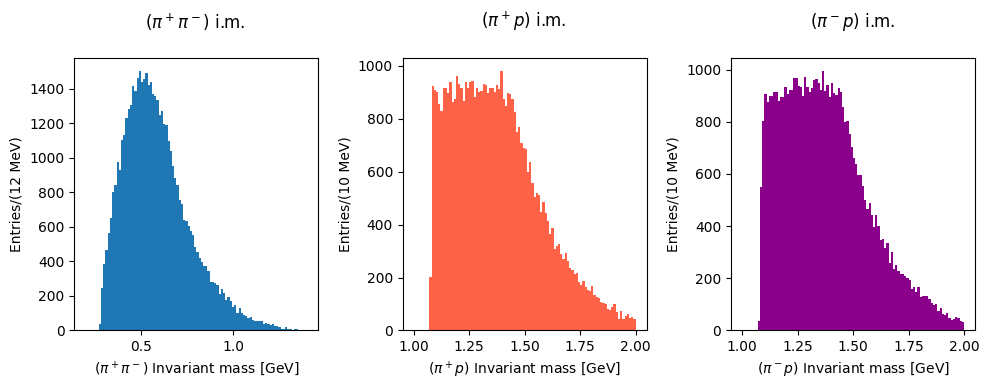
Invariant mass distributions#
Measured data#
Exercise 9
Evaluate the invariant masses (squared and linear) for particle pairs, in a similar way as done in the first example.
Solution to Exercise 9
# @title Solution
inv_mass_squared_12 = (
(data.E1 + data.E2) ** 2
- (data.p1x + data.p2x) ** 2
- (data.p1y + data.p2y) ** 2
- (data.p1z + data.p2z) ** 2
)
inv_mass_squared_13 = (
(data.E1 + data.E3) ** 2
- (data.p1x + data.p3x) ** 2
- (data.p1y + data.p3y) ** 2
- (data.p1z + data.p3z) ** 2
)
inv_mass_squared_23 = (
(data.E3 + data.E2) ** 2
- (data.p3x + data.p2x) ** 2
- (data.p3y + data.p2y) ** 2
- (data.p3z + data.p2z) ** 2
)
invariant_mass_12 = np.sqrt(inv_mass_squared_12)
invariant_mass_13 = np.sqrt(inv_mass_squared_13)
invariant_mass_23 = np.sqrt(inv_mass_squared_23)
Exercise 10
Plot the evaluated invariant masses. First, though, let’s plot the antineutron momentum to see how the distribution looks like.
Solution to Exercise 10
Exercise 11
Now plot the invariant masses distributions for the three pion pairs.
Solution to Exercise 11
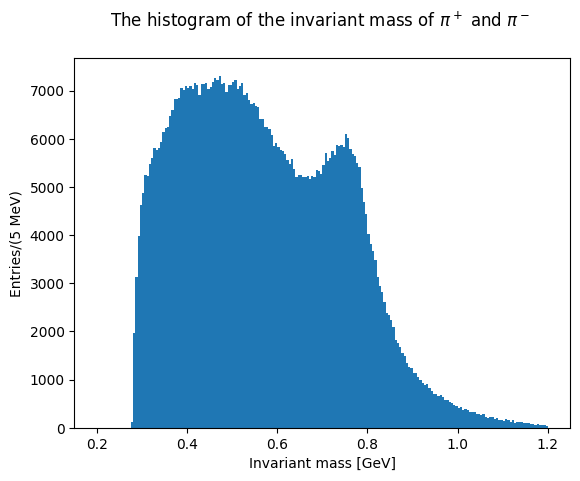
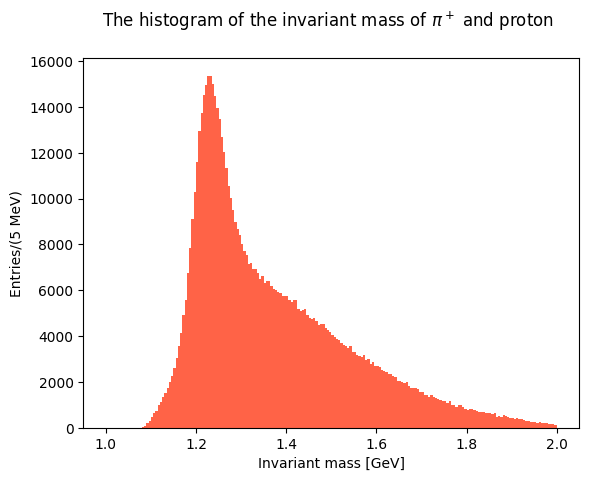
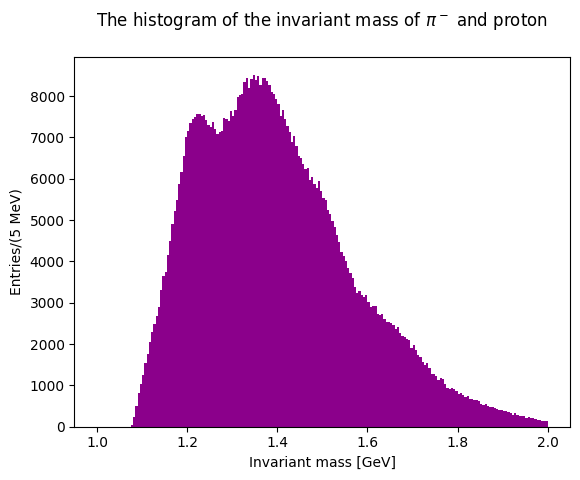
You can superimpose the two last plots to check the differences in the lineshapes of the two distributions.
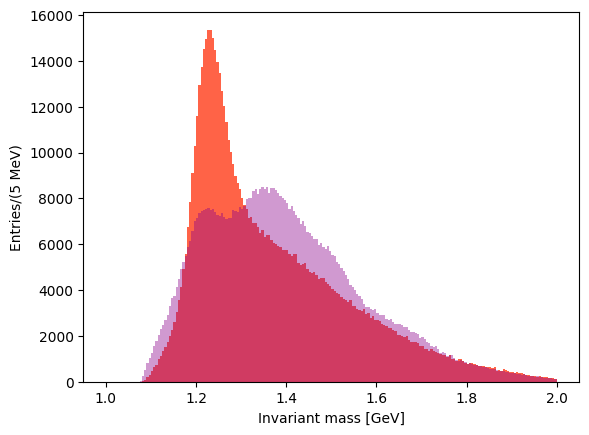
Exercise 12
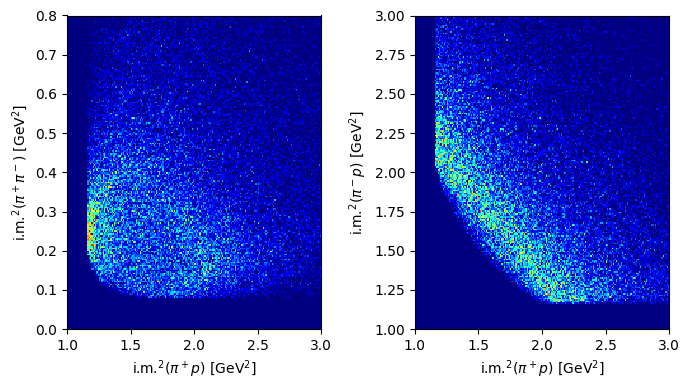
Plot the distributions over the Dalitz plane.
Solution to Exercise 12
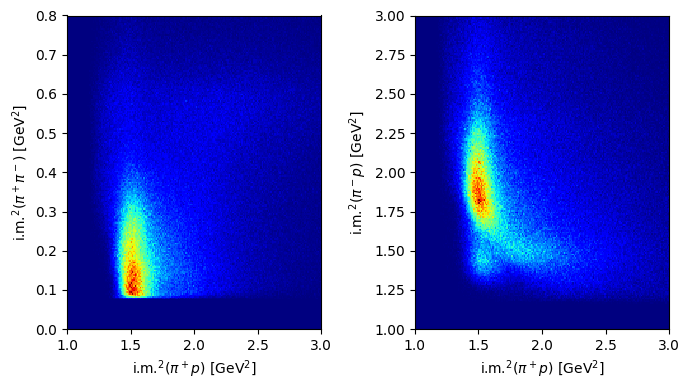
Monte Carlo data#
How do Dalitz plots look like with MonteCarlo generated data? Repeat the previous procedures with a new file, corresponding to generated data from the same reaction, and compare the shapes (statistics are different)
/home/runner/work/strong2020-salamanca/strong2020-salamanca/docs/lecture12a-gammap-solution/.venv/lib/python3.13/site-packages/gdown/cached_download.py:102: FutureWarning: md5 is deprecated in favor of hash. Please use hash='md5:xxx...' instead.
warnings.warn(
# @title Solution
inv_mass_squared_12_mc = (
(mc.E1 + mc.E2) ** 2
- (mc.p1x + mc.p2x) ** 2
- (mc.p1y + mc.p2y) ** 2
- (mc.p1z + mc.p2z) ** 2
)
inv_mass_squared_13_mc = (
(mc.E1 + mc.E3) ** 2
- (mc.p1x + mc.p3x) ** 2
- (mc.p1y + mc.p3y) ** 2
- (mc.p1z + mc.p3z) ** 2
)
inv_mass_squared_23_mc = (
(mc.E3 + mc.E2) ** 2
- (mc.p3x + mc.p2x) ** 2
- (mc.p3y + mc.p2y) ** 2
- (mc.p3z + mc.p2z) ** 2
)
The phase space simulation was done extracting the photon momentum from the real data distribution.
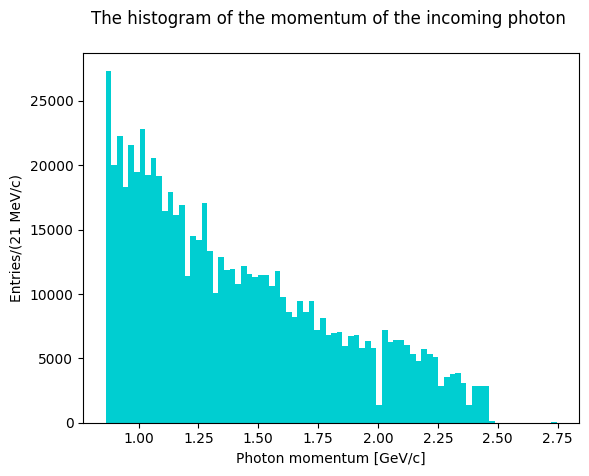
Let’s see the invariant masses histogram shapes using phase space Monte Carlo events.
And now the Dalitz plots:
Missing mass distributions#
Now, let’s take a look to the missing mass distribution (final - initial
state). Let’s start from the real data using the pylorentz package to build 4-vectors (first, work out the lecture12b-kinematics/index.ipynb notebook).
# final state
pip = Momentum4(data.E1, data.p1x, data.p1y, data.p1z)
pim = Momentum4(data.E2, data.p2x, data.p2y, data.p2z)
proton = Momentum4(data.E3, data.p3x, data.p3y, data.p3z)
# initial state
m_proton = 0.93827
n_events = len(data)
zeros = np.zeros(n_events)
ones = np.ones(n_events)
gamma = Momentum4(data.pgamma, zeros, zeros, data.pgamma)
target = Momentum4(m_proton * ones, zeros, zeros, zeros)
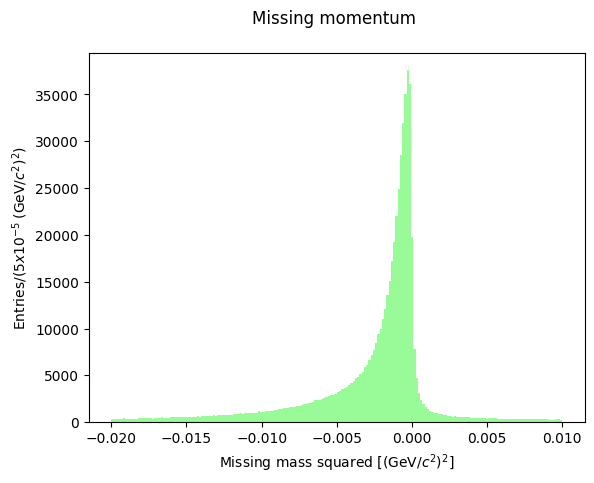
The missing mass square is always negative: this means that the total energy of the initial state exceeds the measured energy of the final state, so there is (likely) a missing particle which carries away some energy. We know that the reaction occurred on a HD molecule as a target: this means that a recoiling neutron is present in all cases when the reaction occurred on a deuteron nucleus: \(\gamma p(n)\rightarrow \pi^+\pi^- p (n)\). In this case, moreover, the hit proton is not at rest but it may have a momentum (called Fermi momentum) which, in the deuteron center-of-mass, is roughly distributed as a gaussian 50 MeV/c wide, with maximum at abouth 270 MeV/c. The missing mass momentum distribution shows the effect of the presence of a non-null Fermi momentum, and the possible contribution to the reaction kinematics of the spectator neutron.
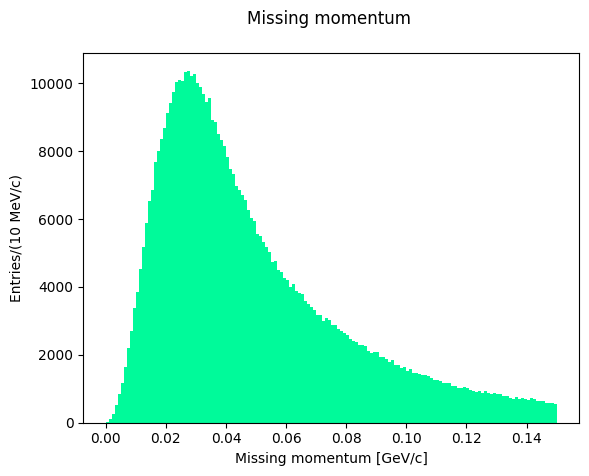
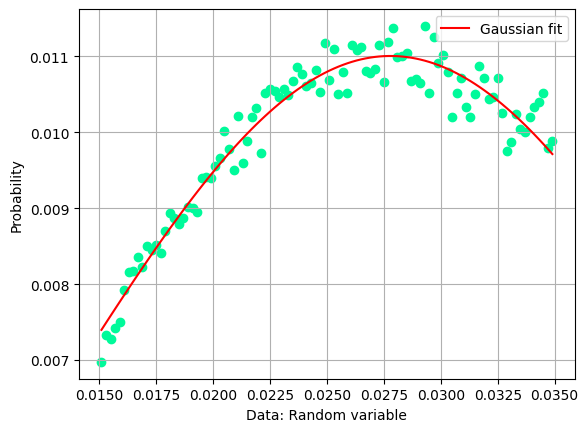
In the data selection procedure, the Fermi momentum was required not to exceed 100 MeV/c to preserve the condition that the neutron is a spectator in the reaction occurring on deuteron. Let’s see the shape of the missing momentum distribution:
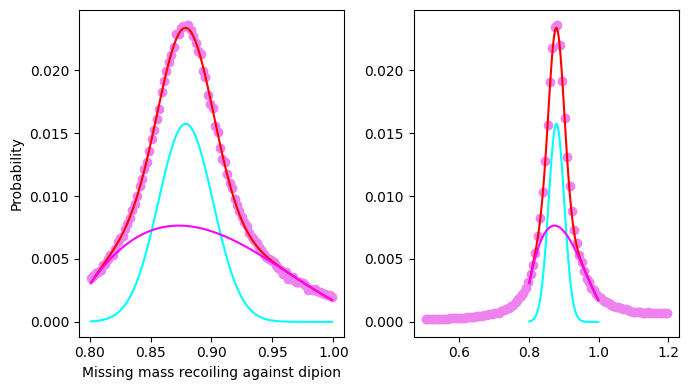
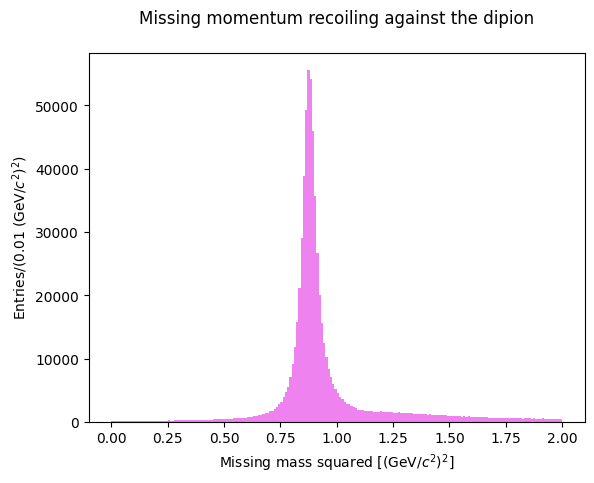
Let’s now consider the missing mass recoiling against the neutral dipion: in an exclusive reaction we expect it to peak at the proton mass. Is the PID selection of our sample acceptable?
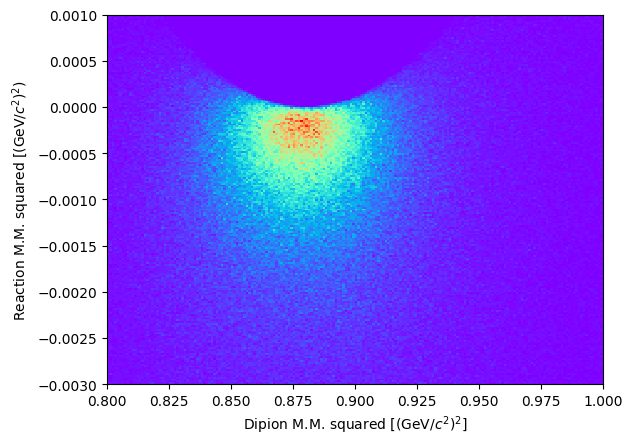
Let’s visualize the scatter plot of the two missing masses squared:
Let’s do some fits#
Which is the maximum of the missing momentum, and where is the peak of the missing mass distribution? Let’s attempt two fits with a single gaussian function.
Fit parameters:
=====================================================
C = 0.011005709742475958 +- 3.732733546918441e-05
X_mean = 0.027786976745429046 +- 0.0001290654386390183
sigma = 0.014235101532634931 +- 0.0002582282886524801
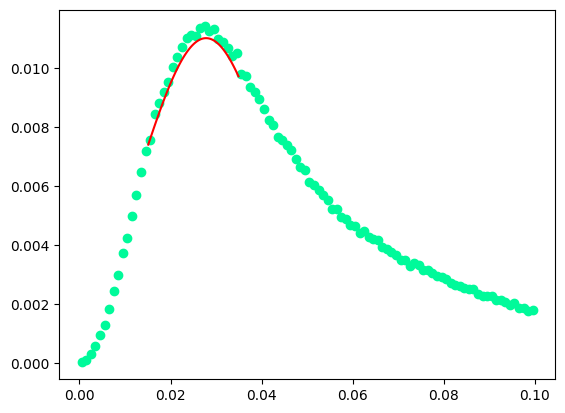
Fit parameters:
=====================================================
C = 0.01142578646237564 +- 7.980412036376522e-05
X_mean = -0.000298904676531158 +- 1.441774961552839e-05
sigma = 0.0006508651288804423 +- 2.1985903713378084e-05
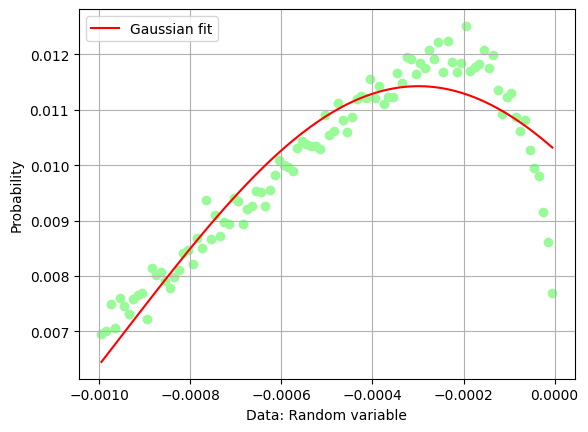
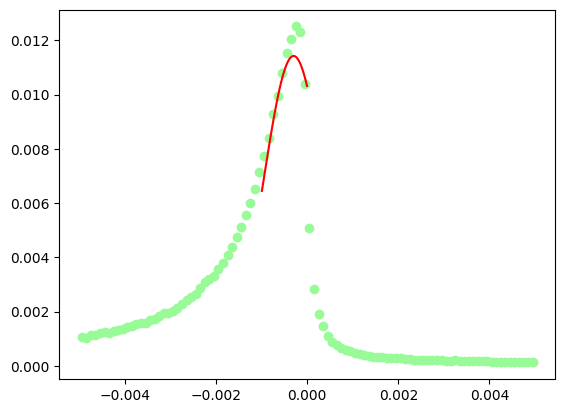
The fit of the missing mass peak is not really very good, one should add some sort of background on the left hand side of the peak, say a 3rd degree polynomial
Fit parameters:
=====================================================
C = 0.012374654548228295 +- inf
X_mean = -0.0003200492404461684 +- inf
sigma = 2.564970506311385e-08 +- inf
missing mass at the peak 0.017889920079367832 GeV/c2
/tmp/ipykernel_2667/2286499957.py:34: OptimizeWarning: Covariance of the parameters could not be estimated
param_optimised, param_covariance_matrix = curve_fit(
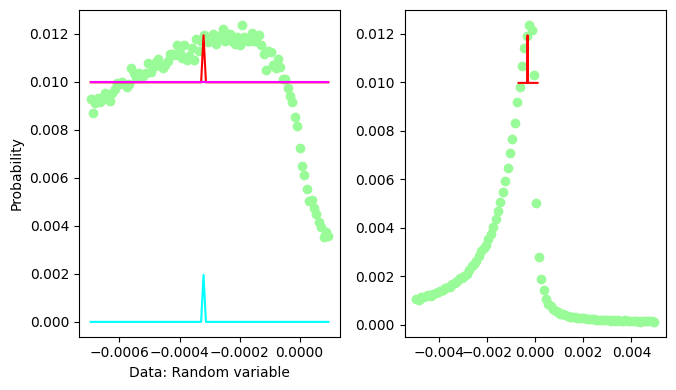
It looks like the Gaussian contribution is refused by the fit.
Let’s make a similar fit for the missing mass recoiling against the dipion:
Fit parameters:
=====================================================
C = 0.015750268159077135 +- 0.00020064765153469743
X_mean = 0.8788503348892618 +- 0.00014610905735201504
sigma = 0.02245275891529404 +- 0.0002493798099026847
missing mass at the peak 0.9374701781332896 GeV/c2