Amplitude model with ampform#
Formulate helicity amplitude model for \(p \gamma \to \eta \pi^0 p\) symbolically using AmpForm.
Generate transitions#
pgamma1 = Particle(
name="pgamma1",
latex=r"p\gamma (s1/2)",
spin=0.5,
mass=4.101931071854584,
charge=1,
isospin=Spin(1 / 2, +1 / 2),
baryon_number=1,
parity=-1,
pid=99990,
)
pgamma2 = create_particle(
template_particle=pgamma1,
name="pgamma2",
latex=R"p\gamma (s3/2)",
spin=1.5,
pid=pgamma1.pid + 1,
)
particle_db.update([pgamma1, pgamma2])
We keep the relevant information about branching fractions in the previous Branching fraction chapter.
For simplicity, after the information about branching fraction, and under the current qrules results, we limit the intermediate resonances to be:
\(a(2)\) for \(\eta \pi^0\),
\(\Delta(1232)\) for \(\pi p\)”,
and “\(N(1535)\)” for both \(\pi^0 p\) and \(\eta p\).
reaction = qrules.generate_transitions(
initial_state="pgamma1",
final_state=["eta", "pi0", "p"],
allowed_intermediate_particles=["a(2)(1320)", "N(1535)", "Delta(1232)"],
allowed_interaction_types=["strong", "EM"],
particle_db=particle_db,
max_angular_momentum=3,
max_spin_magnitude=3,
mass_conservation_factor=0,
)
Formulate model#
model_builder = ampform.get_builder(reaction)
model_builder.config.scalar_initial_state_mass = True
model_builder.config.stable_final_state_ids = 0, 1, 2
bw_builder = RelativisticBreitWignerBuilder(
energy_dependent_width=False,
form_factor=False,
)
for name in reaction.get_intermediate_particles().names:
model_builder.dynamics.assign(name, bw_builder)
model = model_builder.formulate()
The first component of the full amplitude is shown below. The other terms are in similar way of formualation and thus not showing explicitly here again.
\[\begin{split}\displaystyle \begin{aligned}
A^{01}_{- \frac{1}{2}, 0, 0, - \frac{1}{2}} \;&=\; \frac{C_{p\gamma (s1/2) \xrightarrow[S=3/2]{L=1} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} C^{0,0}_{0,0,0,0} C^{\frac{1}{2},- \frac{1}{2}}_{1,0,\frac{3}{2},- \frac{1}{2}} C^{\frac{3}{2},- \frac{1}{2}}_{2,-1,\frac{1}{2},\frac{1}{2}} C^{2,0}_{2,0,0,0} D^{\frac{1}{2}}_{- \frac{1}{2},- \frac{1}{2}}\left(- \phi_{01},\theta_{01},0\right) D^{2}_{-1,0}\left(- \phi^{01}_{0},\theta^{01}_{0},0\right)}{- i \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} - m_{01}^{2} + \left(m_{a_{2}(1320)^{0}}\right)^{2}} \\
\;&+\; \frac{C_{p\gamma (s1/2) \xrightarrow[S=3/2]{L=1} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} C^{0,0}_{0,0,0,0} C^{\frac{1}{2},\frac{1}{2}}_{1,0,\frac{3}{2},\frac{1}{2}} C^{2,0}_{2,0,0,0} C^{\frac{3}{2},\frac{1}{2}}_{2,0,\frac{1}{2},\frac{1}{2}} D^{\frac{1}{2}}_{- \frac{1}{2},\frac{1}{2}}\left(- \phi_{01},\theta_{01},0\right) D^{2}_{0,0}\left(- \phi^{01}_{0},\theta^{01}_{0},0\right)}{- i \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} - m_{01}^{2} + \left(m_{a_{2}(1320)^{0}}\right)^{2}} \\
\;&+\; \frac{C_{p\gamma (s1/2) \xrightarrow[S=5/2]{L=3} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} C^{0,0}_{0,0,0,0} C^{\frac{5}{2},- \frac{1}{2}}_{2,-1,\frac{1}{2},\frac{1}{2}} C^{2,0}_{2,0,0,0} C^{\frac{1}{2},- \frac{1}{2}}_{3,0,\frac{5}{2},- \frac{1}{2}} D^{\frac{1}{2}}_{- \frac{1}{2},- \frac{1}{2}}\left(- \phi_{01},\theta_{01},0\right) D^{2}_{-1,0}\left(- \phi^{01}_{0},\theta^{01}_{0},0\right)}{- i \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} - m_{01}^{2} + \left(m_{a_{2}(1320)^{0}}\right)^{2}} \\
\;&+\; \frac{C_{p\gamma (s1/2) \xrightarrow[S=5/2]{L=3} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} C^{0,0}_{0,0,0,0} C^{2,0}_{2,0,0,0} C^{\frac{5}{2},\frac{1}{2}}_{2,0,\frac{1}{2},\frac{1}{2}} C^{\frac{1}{2},\frac{1}{2}}_{3,0,\frac{5}{2},\frac{1}{2}} D^{\frac{1}{2}}_{- \frac{1}{2},\frac{1}{2}}\left(- \phi_{01},\theta_{01},0\right) D^{2}_{0,0}\left(- \phi^{01}_{0},\theta^{01}_{0},0\right)}{- i \Gamma_{a_{2}(1320)^{0}} m_{a_{2}(1320)^{0}} - m_{01}^{2} + \left(m_{a_{2}(1320)^{0}}\right)^{2}} \\
\end{aligned}\end{split}\]
\[\begin{split}\displaystyle \begin{aligned}
C_{p\gamma (s1/2) \xrightarrow[S=1/2]{L=1} N(1535)^{+} \eta; N(1535)^{+} \xrightarrow[S=1/2]{L=0} p \pi^{0}} \;&=\; 1+0i \\
C_{p\gamma (s1/2) \xrightarrow[S=1/2]{L=1} N(1535)^{+} \pi^{0}; N(1535)^{+} \xrightarrow[S=1/2]{L=0} \eta p} \;&=\; 1+0i \\
C_{p\gamma (s1/2) \xrightarrow[S=3/2]{L=1} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \;&=\; 1+0i \\
C_{p\gamma (s1/2) \xrightarrow[S=3/2]{L=2} \Delta(1232)^{+} \eta; \Delta(1232)^{+} \xrightarrow[S=1/2]{L=1} p \pi^{0}} \;&=\; 1+0i \\
C_{p\gamma (s1/2) \xrightarrow[S=5/2]{L=3} a_{2}(1320)^{0} p; a_{2}(1320)^{0} \xrightarrow[S=0]{L=2} \eta \pi^{0}} \;&=\; 1+0i \\
\Gamma_{N(1535)^{+}} \;&=\; 0.15 \\
\Gamma_{\Delta(1232)^{+}} \;&=\; 0.117 \\
\Gamma_{a_{2}(1320)^{0}} \;&=\; 0.107 \\
m_{0} \;&=\; 0.547862 \\
m_{012} \;&=\; 4.101931071854584 \\
m_{1} \;&=\; 0.1349768 \\
m_{2} \;&=\; 0.93827208816 \\
m_{N(1535)^{+}} \;&=\; 1.53 \\
m_{\Delta(1232)^{+}} \;&=\; 1.232 \\
m_{a_{2}(1320)^{0}} \;&=\; 1.3182 \\
\end{aligned}\end{split}\]
\[\begin{split}\displaystyle \begin{aligned}
m_{01} \;&=\; m_{{p}_{01}} \\
m_{02} \;&=\; m_{{p}_{02}} \\
m_{12} \;&=\; m_{{p}_{12}} \\
\phi_{0} \;&=\; \phi\left({p}_{12}\right) \\
\phi^{01}_{0} \;&=\; \phi\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{01}}\right|}{E\left({p}_{01}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{01}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{01}\right)\right) p_{0}\right) \\
\phi^{02}_{0} \;&=\; \phi\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{02}}\right|}{E\left({p}_{02}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{02}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{02}\right)\right) p_{0}\right) \\
\phi_{01} \;&=\; \phi\left({p}_{01}\right) \\
\phi^{12}_{1} \;&=\; \phi\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{12}}\right|}{E\left({p}_{12}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{12}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{12}\right)\right) p_{1}\right) \\
\phi_{02} \;&=\; \phi\left({p}_{02}\right) \\
\theta_{0} \;&=\; \theta\left({p}_{12}\right) \\
\theta^{01}_{0} \;&=\; \theta\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{01}}\right|}{E\left({p}_{01}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{01}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{01}\right)\right) p_{0}\right) \\
\theta^{02}_{0} \;&=\; \theta\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{02}}\right|}{E\left({p}_{02}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{02}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{02}\right)\right) p_{0}\right) \\
\theta_{01} \;&=\; \theta\left({p}_{01}\right) \\
\theta^{12}_{1} \;&=\; \theta\left(\boldsymbol{B_z}\left(\frac{\left|\vec{{p}_{12}}\right|}{E\left({p}_{12}\right)}\right) \boldsymbol{R_y}\left(- \theta\left({p}_{12}\right)\right) \boldsymbol{R_z}\left(- \phi\left({p}_{12}\right)\right) p_{1}\right) \\
\theta_{02} \;&=\; \theta\left({p}_{02}\right) \\
\end{aligned}\end{split}\]
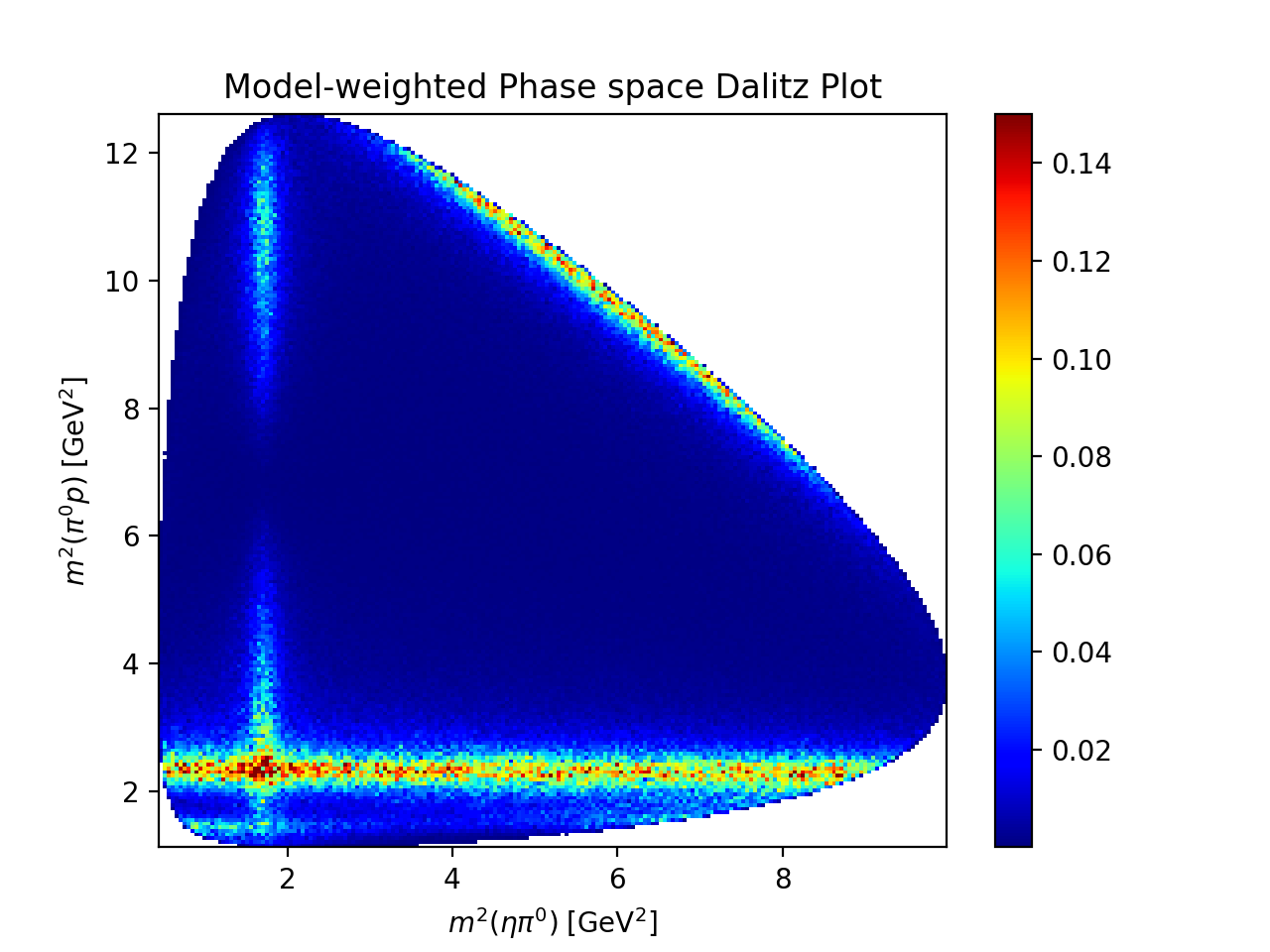
Visualization#
unfolded_expression = model.expression.doit()
intensity_func = create_parametrized_function(
expression=unfolded_expression,
parameters=model.parameter_defaults,
backend="jax",
)
phsp_event = 500_000
rng = TFUniformRealNumberGenerator(seed=0)
phsp_generator = TFPhaseSpaceGenerator(
initial_state_mass=reaction.initial_state[-1].mass,
final_state_masses={i: p.mass for i, p in reaction.final_state.items()},
)
phsp_momenta = phsp_generator.generate(phsp_event, rng)
helicity_transformer = SympyDataTransformer.from_sympy(
model.kinematic_variables,
backend="jax",
)
phsp = helicity_transformer(phsp_momenta)
{0: ['Delta(1232)+', 'N(1535)+'], 1: ['N(1535)+'], 2: ['a(2)(1320)0']}
%matplotlib widget