Amplitude model with ampform#
PWA study on \(p \gamma \to \Lambda K^+ \pi^0\).
We formulate the helicity amplitude model symbolically using AmpForm here.
Decay definition#
Particle definitions#
Particle |
Name |
PID |
\(J^{PC} (I^G)\) |
\(I_3\) |
\(M\) |
\(\Gamma\) |
\(Q\) |
\(S\) |
\(B\) |
|---|---|---|---|---|---|---|---|---|---|
\(\Lambda\) |
|
3122 |
\(\frac{1}{2}^{+ } \; (0^{ })\) |
\(0\) |
1.12 |
2.515e-15 |
0 |
-1 |
1 |
\(K^{+}\) |
|
321 |
\(0^{- } \; (\frac{1}{2}^{ })\) |
\(\frac{1}{2}\) |
0.494 |
5.317e-17 |
1 |
1 |
0 |
\(\pi^{0}\) |
|
111 |
\(0^{-+} \; (1^{-})\) |
\(0\) |
0.135 |
7.81e-09 |
0 |
0 |
0 |
\(\gamma\) |
|
22 |
\(1^{--}\) |
N/A |
0 |
0 |
0 |
0 |
0 |
\(p\) |
|
2212 |
\(\frac{1}{2}^{+ } \; (\frac{1}{2}^{ })\) |
\(\frac{1}{2}\) |
0.938 |
0 |
1 |
0 |
1 |
In the table above, PID is the PDG ID from PDG particle numbering scheme, \(J\) is the spin, \(P\) is the parity, \(C\) is the C parity, \(I\) is the isospin (magnitude), \(G\) is the G parity. \(I_3\) is the isospin projection (or the 3rd component), \(M\) is the mass, \(\Gamma\) is the width, \(Q\) is the charge, \(S\) is the strangeness number, and \(B\) is the baryon number.
Initial state definition#
Mass for \(p \gamma\) system
E_lab_gamma = 8.5
m_proton = 0.938
m_0 = np.sqrt(2 * E_lab_gamma * m_proton + m_proton**2)
m_eta = 0.548
m_pi = 0.135
m_0
np.float64(4.101931740046389)
Add custom particle \(p \gamma\)
pgamma1 = Particle(
name="pgamma1",
latex=r"p\gamma (s1/2)",
spin=0.5,
mass=m_0,
charge=1,
isospin=Spin(1 / 2, +1 / 2),
baryon_number=1,
parity=-1,
pid=99990,
)
pgamma2 = create_particle(
template_particle=pgamma1,
name="pgamma2",
latex=R"p\gamma (s3/2)",
spin=1.5,
pid=pgamma1.pid + 1,
)
particle_db.update([pgamma1, pgamma2])
Generate transitions#
For simplicity, we use the initial state \(p \gamma\) (with spin-\(\frac{1}{2}\)), and set the allowed interaction type to be strong only, the formalism is selected to be helicity formalism instead of canonical.
See also
reaction = qrules.generate_transitions(
initial_state=("pgamma1"),
final_state=["Lambda", "K+", "pi0"],
allowed_interaction_types=["strong"],
formalism="helicity",
particle_db=particle_db,
max_angular_momentum=4,
max_spin_magnitude=4,
mass_conservation_factor=0,
)
Formulate amplitude model#
model_builder = ampform.get_builder(reaction)
model_builder.config.scalar_initial_state_mass = True
model_builder.config.stable_final_state_ids = 0, 1, 2
bw_builder = RelativisticBreitWignerBuilder(
energy_dependent_width=False,
form_factor=False,
)
for name in reaction.get_intermediate_particles().names:
model_builder.dynamics.assign(name, bw_builder)
model = model_builder.formulate()
model.intensity
The first term in the amplitude model:
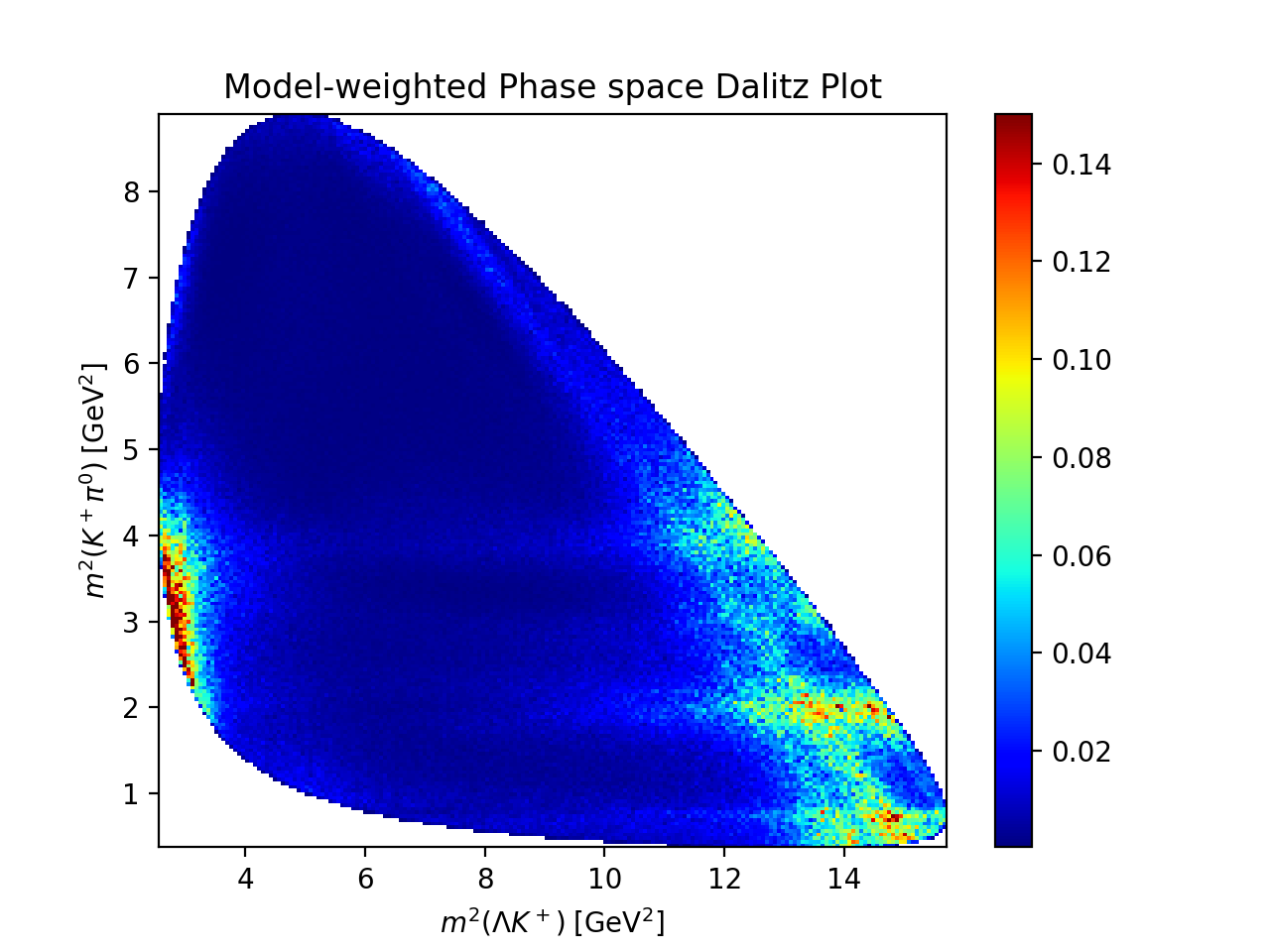
Visualization#
unfolded_expression = model.expression.doit()
intensity_func = create_parametrized_function(
expression=unfolded_expression,
parameters=model.parameter_defaults,
backend="jax",
)
phsp_event = 500_000
rng = TFUniformRealNumberGenerator(seed=0)
phsp_generator = TFPhaseSpaceGenerator(
initial_state_mass=reaction.initial_state[-1].mass,
final_state_masses={i: p.mass for i, p in reaction.final_state.items()},
)
phsp_momenta = phsp_generator.generate(phsp_event, rng)
helicity_transformer = SympyDataTransformer.from_sympy(
model.kinematic_variables,
backend="jax",
)
phsp = helicity_transformer(phsp_momenta)
%matplotlib widget