Phase space for a three-body decay#
Kinematics for a three-body decay \(0 \to 123\) can be fully described by two Mandelstam variables \(\sigma_1, \sigma_2\), because the third variable \(\sigma_3\) can be expressed in terms \(\sigma_1, \sigma_2\), the mass \(m_0\) of the initial state, and the masses \(m_1, m_2, m_3\) of the final state. As can be seen, the roles of \(\sigma_1, \sigma_2, \sigma_3\) are interchangeable.
The phase space is defined by the closed area that satisfies the condition \(\phi(\sigma_1,\sigma_2) \leq 0\), where \(\phi\) is a Kibble function:
and \(\lambda\) is the Källén function:
Any distribution over the phase space can now be defined using a two-dimensional grid over a Mandelstam pair \(\sigma_1,\sigma_2\) of choice, with the condition \(\phi(\sigma_1,\sigma_2)<0\) selecting the values that are physically allowed.
phsp_expr = is_within_phasespace(s1, s2, m0, m1, m2, m3, outside_value=0)
phsp_func = create_parametrized_function(
phsp_expr.doit(),
parameters={m0: 2.2, m1: 0.2, m2: 0.4, m3: 0.4},
backend="numpy",
)
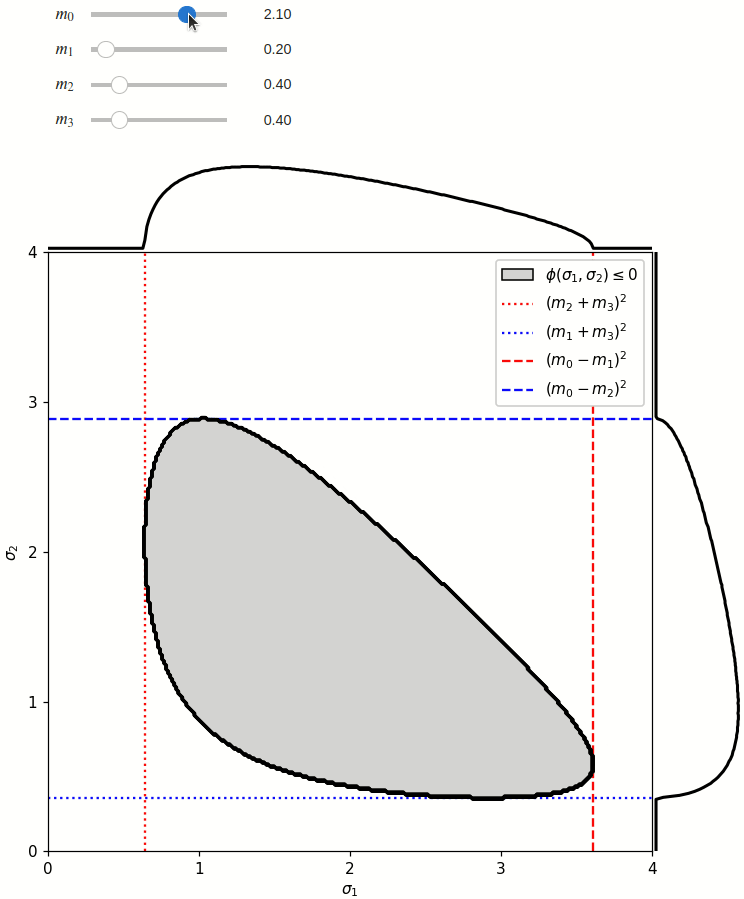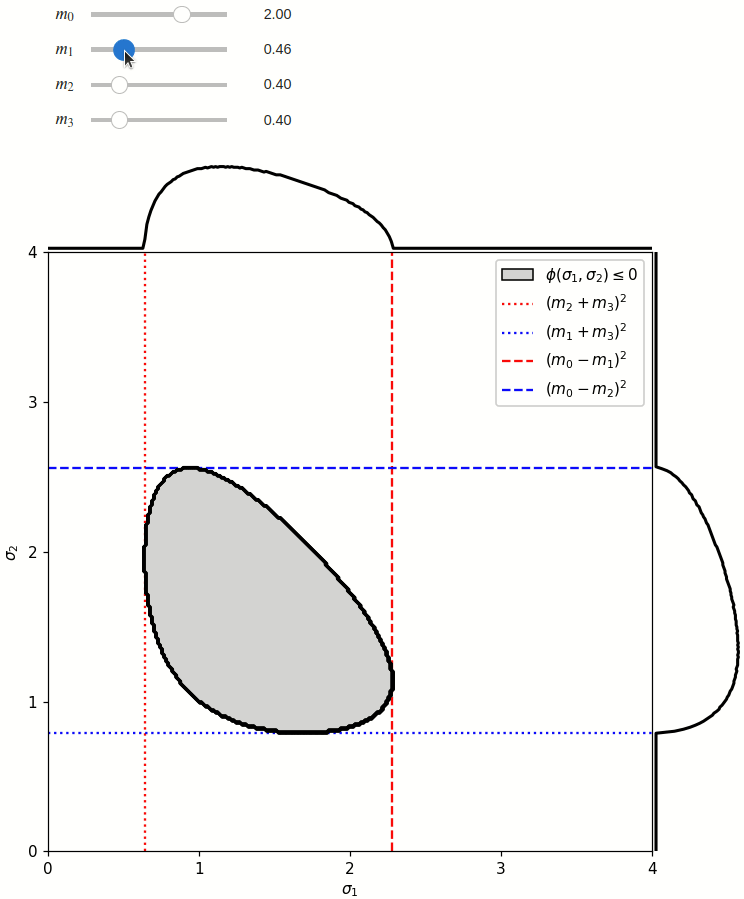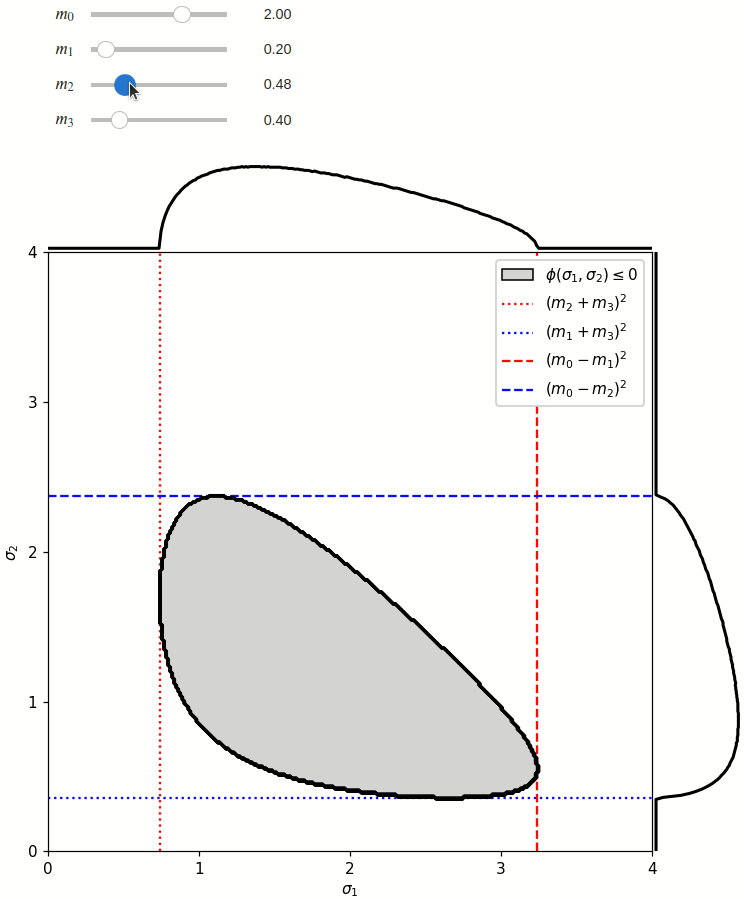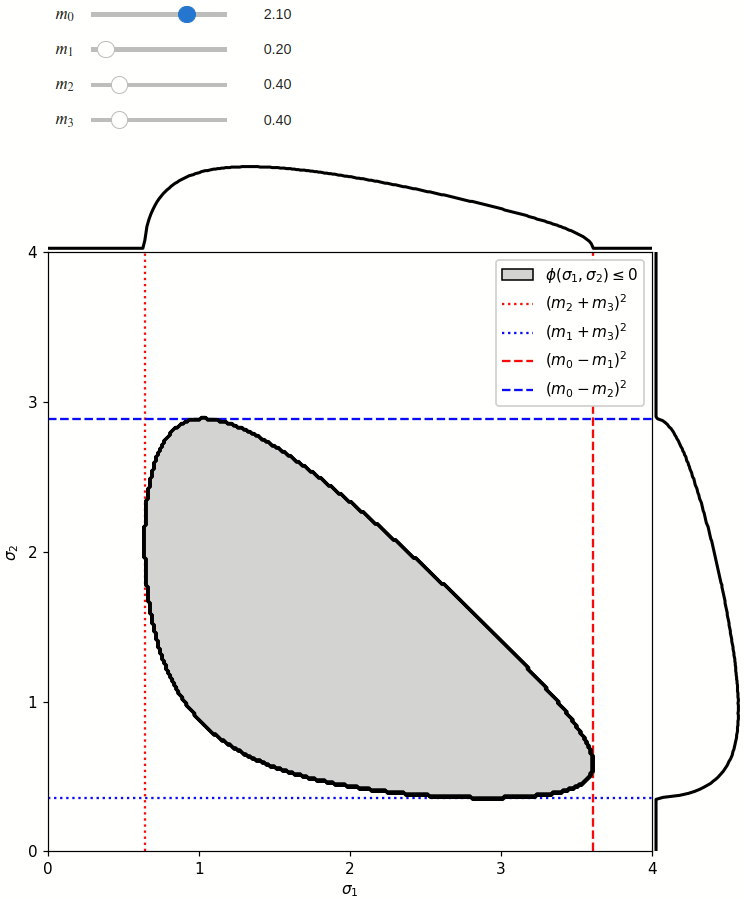
The phase space boundary can be described analytically in terms of \(\sigma_1\) or \(\sigma_2\), in which case there are two solutions:
sol1, sol2 = sp.solve(kibble.doit().subs(s3, computed_s3), s2)
The boundary cannot be parametrized analytically in polar coordinates, but there is a numeric solution. The idea is to solve the condition \(\phi(\sigma_1,\sigma_2)=0\) after the following substitutions:
For every value of \(\theta \in [0, 2\pi)\), the value of \(r\) can now be found by solving the condition \(\phi(r, \theta)=0\). Note that \(\phi(r, \theta)\) is a cubic polynomial of \(r\). For instance, if we take \(m_0=5, m_1=2, m_{2,3}=1\):
The lowest value of \(r\) that satisfies \(\phi(r,\theta)=0\) defines the phase space boundary.



